Parkinson’s disease (PD) is the second most common neurodegenerative disease characterized by motor disturbances such as resting tremor, rigidity, postural instability, gait problems, and gastrointestinal dysfunction. Recent studies suggest that alterations in the gut phagobiota may contribute to PD. Our analyses for the first time revealed significant alterations in the representation of certain bacteriophages in the phagobiota of PD patients. We identified shifts of the phage/bacteria ratio in lactic acid bacteria known to produce dopamine and regulate intestinal permeability, which are major factors implicated in PD pathogenesis. Furthermore, we observed the depletion of Lactococcus spp. in the PD group, which was most likely due to the increase of lytic c2-like and 936-like lactococcal phages frequently present in dairy products. Our findings add bacteriophages to the list of possible factors associated with the development of PD, suggesting that gut phagobiota composition may serve as a diagnostic tool as well as a target for therapeutic intervention, which should be confirmed in further studies. Our results open a discussion on the role of environmental phages and phagobiota composition in health and disease.
Parkinson’s Disease and Bacteriophages
We discovered striking alterations in the abundance of strictly lytic bacteriophages in patients with very early-stage Parkinson’s disease that most likely contributed to the disease onset.
Parkinson’s disease and bacteriophages as its contributors:
Propagation of α-synucleinmisfolding

HMI was the first to identify various prion-like domains that have a strong capability to become prions in multiple bacteriophages, including those associated with the human microbiota. The involvement of these bacteriophage prion domains in cross-kingdom interactions with eukaryotic proteins and in α-synucleinmisfolding is currently under investigation by HMI.
Phage-Induced Alterations in the Abundances of Lactococcus spp. Might Trigger Parkinson’s Disease
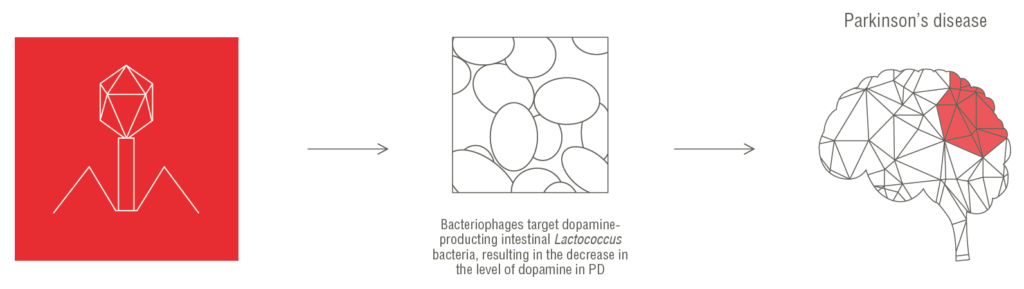
HMI revealed significant alterations in the abundances of certain bacteriophages in the phagobiota of patients with Parkinson’s disease. We identified shifts in the phage/bacteria ratio of Lactococcus spp., which is a lactic acid bacterium that is known to produce dopamine and regulate intestinal permeability, which are major factors implicated in the pathogenesis of Parkinson’s disease. Furthermore, we observed the depletion of Lactococcus spp. in the group with Parkinson’s disease, which was most likely caused by an increased abundance of lytic c2-like and 936-like lactococcal phages. Our findings add bacteriophages to the list of possible factors associated with the development of Parkinson’s disease, suggesting that the gut phagobiota composition may serve as a diagnostic tool as well as a target for therapeutic intervention.
Bacteriophages Induce Elevated Levels of PAMPs
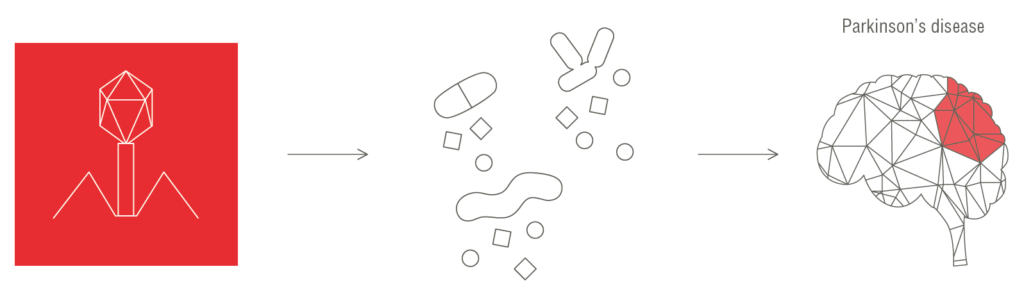
We showed that bacteriophages can induce increased levels of plasma cell-free bacterial DNA and other PAMPs, such as lipopolysaccharides, peptidoglycan, and bacterial amyloid. These PAMPs can enter the blood circulation, leading to the establishment of chronic inflammation and an altered immune response, both of which are known to be associated with the development and progression of Parkinson’s disease.
The phage-induced increase in intestinal permeability
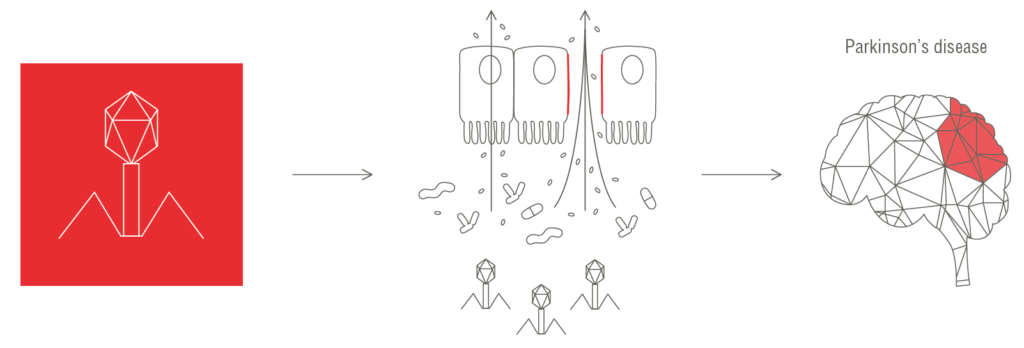
We discovered that certain bacteriophages increase intestinal permeability, facilitating the translocation of gut bacteria and leading to chronic inflammation and an altered immune response, both of which are conditions that are associated with Parkinson’s disease.
Bacteriophages as New Human Viral Pathogens.Microorganisms. (2018); 6.
Microorganisms MDPI (2018)
Bacteriophages as New Human Viral Pathogens
| Publication Type |
Journal article |
| Authors |
George V. Tetz
Victor Tetz |
| Abstract |
The pathogenesis of numerous human multifaceted devastating diseases, including a
variety of neurodegenerative and autoimmune diseases, is associated with alterations in the gut
microbiota; however, the underlying mechanisms are not completely understood. Our recent human metagenome and phagobiota proteome analyses and studies in relevant animal models suggested that bacterial viruses might be implicated in the progression and maintenance of at least some pathologies, including those associated with protein misfolding. Here, for the first time, we propose the concept of bacteriophages as human pathogens. We suggest that bacterial viruses have different
ways to directly and indirectly interact with eukaryotic cells and proteins, leading to human diseases. Furthermore, we suggest different causes of bacteriophages infection on the basis of the unique ways of interplay of phages, microbiota, and the human host. This concept opens a discussion of the role of bacteriophages as previously overlooked pathogenic factors and suggests that bacterial viruses have to be further explored as a diagnostic and treatment target for therapeutic intervention. |
Year of
Publication |
2018 |
| Journal |
Microorganisms MDPI |
| DOI |
10.3390/microorganisms6020054 |
Prion-like Domains in Eukaryotic Viruses.Scientific reports (2018); 12:8931.
Scientific Reports (2018)
Prion-Like Domains in Eukaryotic Viruses
| Publication Type |
Journal article |
| Authors |
George V. Tetz
Victor Tetz |
| Abstract |
Prions are proteins that can self-propagate, leading to the misfolding of proteins. In addition to the previously demonstrated pathogenic roles of prions during the development of different mammalian diseases, including neurodegenerative diseases, they have recently been shown to represent an important functional component in many prokaryotic and eukaryotic organisms and bacteriophages, confirming the previously unexplored important regulatory and functional roles. However, an in-depth analysis of these domains in eukaryotic viruses has not been performed. Here, we examined the presence of prion-like proteins in eukaryotic viruses that play a primary role in different ecosystems and that are associated with emerging diseases in humans. We identified relevant functional associations in different viral processes and regularities in their presence at different taxonomic levels. Using the prion-like amino-acid composition computational algorithm, we detected 2679 unique putative prion-like domains within 2,742,160 publicly available viral protein sequences. Our findings indicate that viral prion-like proteins can be found in different viruses of insects, plants, mammals, and humans. The analysis performed here demonstrated common patterns in the distribution of prion-like domains across viral orders and families, and revealed probable functional associations with different steps of viral replication and interaction with host cells. These data allow the identification of the viral prion-like proteins as potential novel regulators of viral infections. |
Year of
Publication |
2018 |
| Journal |
Scientific Reports |
| DOI |
10.1038/s41598-018-27256-w |
Prion-like domains in phagobiota.Frontiers in microbiology (2017);15:2239.
Frontiers in Microbiology (2017)
Prion-Like Domains in Phagobiota
| Publication Type |
Journal article |
| Authors |
George V. Tetz
Victor Tetz |
| Abstract |
IPrions are molecules characterized by self-propagation, which can undergo a conformational switch leading to the creation of new prions. Prion proteins have originally been associated with the development of mammalian pathologies; however, recently they have been shown to contribute to the environmental adaptation in a variety of prokaryotic and eukaryotic organisms. Bacteriophages are widespread and represent the important regulators of microbiota homeostasis and have been shown to be diverse across various bacterial families. Here, we examined whether bacteriophages contain prion-like proteins and whether these prion-like protein domains are involved in the regulation of homeostasis. We used a computational algorithm, prion-like amino acid composition, to detect prion-like domains in 370,617 publicly available bacteriophage protein sequences, which resulted in the identification of 5040 putative prions. We analyzed a set of these prion-like proteins, and observed regularities in their distribution across different phage families, associated with their interactions with the bacterial host cells. We found that prion-like domains could be found across all phages of various groups of bacteria and archaea. The results obtained in this study indicate that bacteriophage prion-like proteins are predominantly involved in the interactions between bacteriophages and bacterial cell, such as those associated with the attachment and penetration of bacteriophage in the cell, and the release of the phage progeny. These data allow the identification of phage prion-like proteins as novel regulators of the interactions between bacteriophages and bacterial cells. |
Year of
Publication |
2017 |
| Journal |
Frontiers in Microbiology |
| DOI |
10.3389/fmicb.2017.02239 |
Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gutpathogens (2017) 8:33.
Scientific Reports (2017)
Bacteriophages as potential new mammalian pathogens
| Publication Type |
Jornal article |
| Authors |
George V. TetzKelly V. Ruggles
Hua Zhou
Adriana Heguy
Aristotelis Tsirigos
Victor Tetz |
| Abstract |
Increased intestinal permeability and translocation of gut bacteria trigger various polyaetiological diseases associated with chronic inflammation and underlie a variety of poorly treatable pathologies. Previous studies have established a primary role of the microbiota composition and intestinal permeability in such pathologies. Using a rat model, we examined the effects of exposure to a bacteriophage cocktail on intestinal permeability and relative abundance of taxonomic units in the gut bacterial community. There was an increase in markers of impaired gut permeability, such as the lactulose/mannitol ratio, plasma endotoxin concentrations, and serum levels of inflammation-related cytokines, following the bacteriophage challenge. We observed significant differences in the alpha diversity of faecal bacterial species and found that richness and diversity index values increased following the bacteriophage challenge. There was a reduction in the abundance of Blautia, Catenibacterium, Lactobacillus, and Faecalibacterium species and an increase in Butyrivibrio,Oscillospira and Ruminococcus after bacteriophage administration. These findings provide novel insights into the role of bacteriophages as potentially pathogenic for mammals and their possible implication in the development of diseases associated with increased intestinal permeability. |
Year of
Publication |
2017 |
| Journal |
Scientific Reports |
| DOI |
10.10038/s41598-017-07278-6. |
Infection of Microbiota by Bacteriophages Can Be Considered a New Group of Viral Diseases of Mammals Including Humans. ASMMicrobe (2017)
ASM Microbe (2017)
Infection of Microbiota by Bacteriophages Can Be Considered a New Group of Viral Diseases of Mammals Including Humans
| Publication Type |
Oral presentation |
| Authors |
George TetzVictor Tetz |
| Abstract |
Background: The objective of this study was to assess the effect of microbiota treatment with bacteriophages on the intestinal permeability in vivo and to evaluate the possibility that the infection of microbiota by bacteriophages may affect mammals.Methods: We studied alterations in the host macroorganism and increased intestinal permeability as a result of a direct effect of bacteriophage cocktail. Healthy adult, albino Wistar rats, weighing 180-220g were given daily (for 10 days) phage cocktail (1.5 ml of 1×106 plaque forming units/ml) active against Enterobacteriaceae, Staphylococcaceae, Streptococcaceae, and Pseudomonadaceae families (1). The lactulose-mannitol ratio was used as a marker of intestinal permeability (2). Circulating immune complexes (CIC) were evaluated as markers of endogenous intoxication (3). Metagenomic analysis was used to characterize the composition of microbiota before and after phage challenge.Results: After 10 days of challenge, the rats showed weight loss, decreased activity. They displayed a significantly elevated lactulose:mannitol ratio with the mean increase 2.4 fold (2). The level of CIC was more than 2.5 times higher as compared to before treatment, indicating endogenous intoxication, caused by leaky gut (3). Metagenomic analysis revealed phage-induced altered microbiota composition that led to the increased intestinal permeability and transcriptome alterations were relevant to leaky gut syndrome.
Conclusions: To our knowledge, this study for the first time indicates the link between bacteriophages and mammalian pathologies associated with increased intestinal permeability such as systemic inflammatory and psychological or autoimmune disorders. This study demonstrates that increased intestinal permeability may be induced by bacteriophages that affect microbiota. We propose that infection of microbiota by bacteriophages can be considered a new group of viral diseases of mammals including humans (4). |
Year of
Publication |
2017 |
| Journal |
ASM Microbe 2017 |
| DOI |
ASM2017posters. |
Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gutpathogens (2016) 2016; 8:33
Gut pathogens (2016)
Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model
| Publication Type |
Journal Article |
| Authors |
George TetzVictor Tetz |
| Abstract |
Increased intestinal permeability and translocation of gut microbiota from the intestinal lumen to the systemic circulation predispose patients to various diseases and may be one of the main triggers thereof. The role of microbiota in increased intestinal permeability is under intensive investigation. Here, we studied alterations in the host and increased intestinal permeability as a direct effect of treatment with a bacteriophage cocktail. After 10 days of challenge, the rats showed weight loss, messy hair, and decreased activity. Additionally, they displayed a significantly elevated lactulose:mannitol ratio and the level of circulating immune complexes. To our knowledge, this study demonstrates for the first time that increased intestinal permeability may be induced by bacteriophages that affect the microbiota. |
Year of
Publication |
2016 |
| Journal |
Gut pathogens |
| DOI |
10.1186/s13099-016-0109-1 |




